Feature Photo: Pixabay
India is one of the most biologically diverse countries on the planet; it boasts 91,000 animal species, or 8% of known wildlife. The Wildlife Protection Act of 1972 (WPA) officially listed India’s endangered species and designated protections for them in the form of national parks, sanctuaries, and wildlife reserves. Unfortunately, the illegal wildlife trade in India is flourishing, dramatically reducing the populations of many threatened and endangered species. Even after the implementation of the WPA, which also punishes those caught for hunting protected animals, poaching has continued to increase in India. The recent wave of COVID-19 lockdowns led to further increases in the illegal wildlife trade (IWT.) Sporadic border security has contributed to India’s increase in wildlife trading; this, along with the rise in wildlife cybercrimes, has led to India’s rise to an international animal trafficking hub.
Felids, or wild cats, are a common target in the IWT. India is home to 15 of the world’s 36 wild felid species, more than any other nation. Big and small cat species are coveted for their skins, bones, teeth, and claws. Identifying the species being killed and traded, as well as understanding where these body parts originated, is a vital step in tracing the IWT networks and targeting laws and security measures to ensure these species are adequately protected.
Several researchers at the Wildlife Institute of India want to apply the same techniques used to identify humans, such as DNA genotyping, to rapidly and accurately identify poached exotic animals, a booming area in wildlife forensics. Differentiating between wild felid species and identifying which species are most seen in the illegal wildlife trade is vital for drafting legislation that protects these at-risk species. In this study, researchers took hair and tissue samples from the institute’s wildlife forensic laboratory, including fishing cats, jungle cats, caracals, and leopard cats. The researchers aimed to identify unique characteristics in the samples, using microscopic hair analysis as well as mitochondrial DNA analysis, to aid in the identification of wild felid parts seized from traffickers.

Source: Pixabay
Microscopic hair examination is often a preliminary technique in forensic hair analysis. Hair samples have three layers: the outer cuticle, the middle cortex, and the deep medulla. Human and animal hairs noticeably differ in the medulla, akin to the lead of a pencil, which is far wider in animals than humans, and the scale patterns on the cuticle.
The physical characteristics of hair can be a helpful primary step in identifying their source, but merely studying hairs under the microscope cannot reveal their specific origin. The morphological differences among the feline species was not significant enough to use microscopy as the major tool of identification. If a hair sample appears promising, the next step in identification is DNA testing.
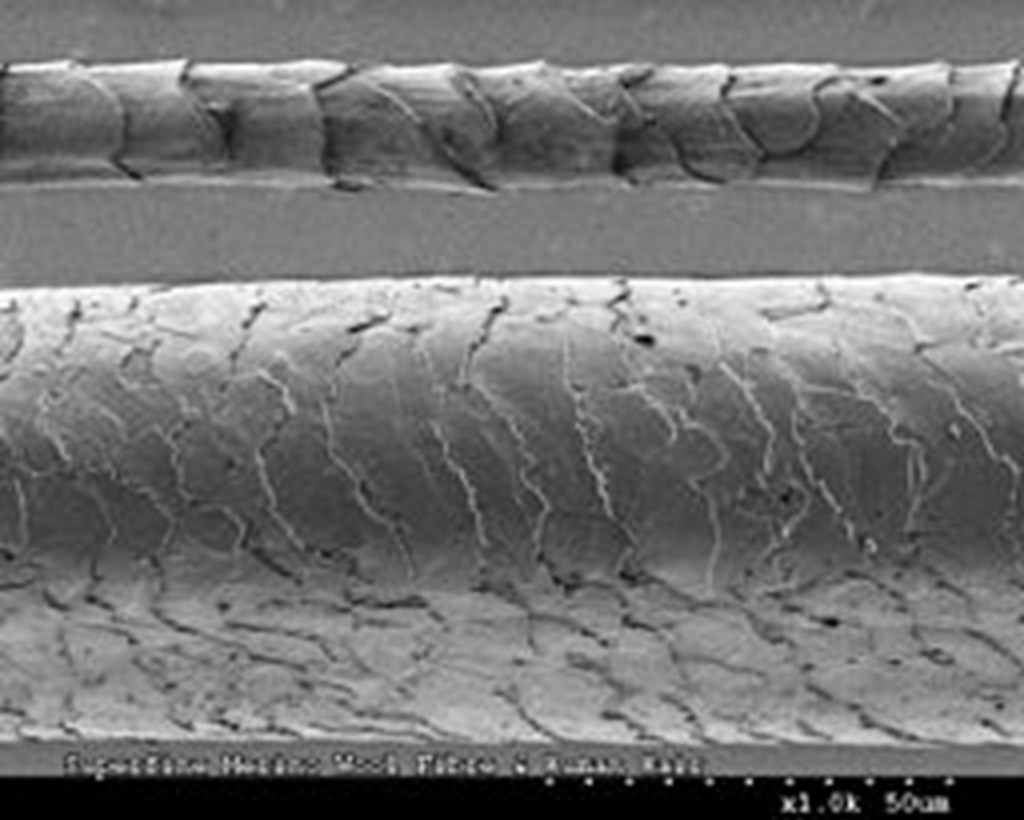
Source: Wikimedia Commons
Mitochondrial DNA, or mtDNA, is a form of DNA that exists in the mitochondria- the powerhouse of the cell. Mitochondrial DNA passes directly from a mother to her children, so it reveals the maternal relatives of a creature, useful in determining evolutionary history. Each mtDNA gene is made up of different combinations of base pairs whose variation signals different familial and evolutionary histories.
In this case, researchers examined mtDNA from five big cat species to determine which portions of mtDNA varied between the species. DNA was extracted from each tissue sample and amplified using Polymerase Chain Reaction, or PCR. PCR can multiply mtDNA with an enzyme, creating an exponential number of copies of the starting material. Researchers then identified three locations on the cats’ mt DNA to study- the 12S rRNA gene, the 16S rRNA gene, and the Cyt b gene. It was important to find genes that could be easily identified in every cat genome but differed enough to show evolutionary variations. These base pair sequences were added to the wildlife DNA database and will serve as the basis for identifying hair samples confiscated from illegal wildlife traders in the future.
Establishing a database of morphological hair characteristics is vital in guiding forensic technicians in identifying illegally traded animal parts, and expanding the mtDNA database provides data to definitively identify the origin of the samples. The forensic examination of hair and tissue samples can allow investigators to identify the species that are most vulnerable to illegal poaching. The addition of hair and tissue samples to the existing database will enable the Wildlife Institute of India to better determine the origin of items intercepted in the fight against illegal poaching.
| Title: Identification of selected wild felids using hair morphology and forensically informative nucleotide sequencing (FINS): Wildlife forensics prospective | |
| Authors: Gagandeep Singh, Yellapu Srinivas, Gandla Chethan Kumar, Ashutosh Singh, Chandra Prakash Sharma, Sandeep Kumar Gupta | |
| Journal: Legal Medicine | |
| Year: 2020 | |
| Link: https://doi.org/10.1016/j.legalmed.2020.101692 |
